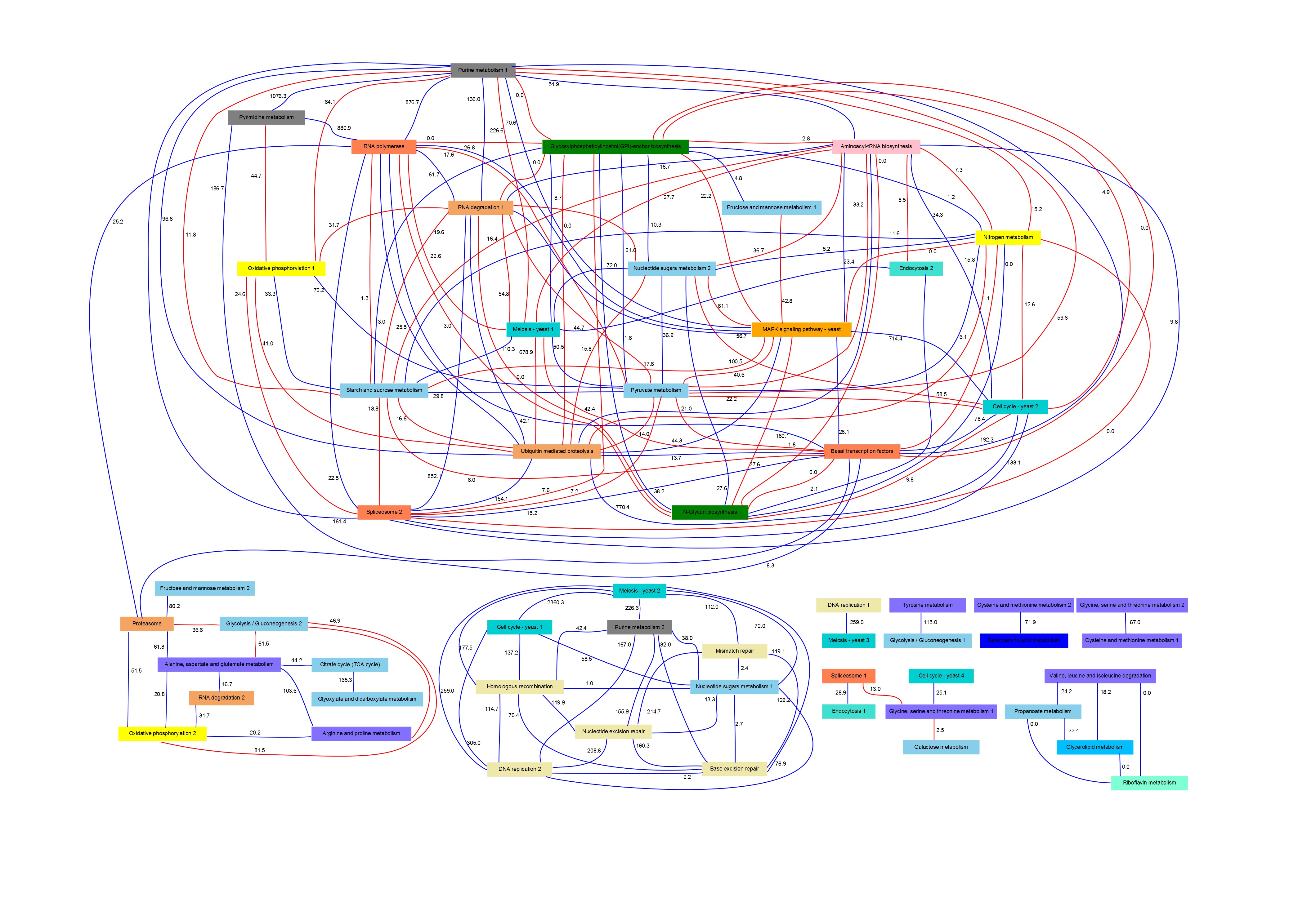
Yeast Pathway Network
PANA was validated using the cdc15 time series published by Spellman et al. (1998), and for the functional annotation of genes into pathways KEGG database was used. The network can by visualized following this link.
The nodes in the network denote pathway signatures and the links show the associations inferred by PANA.
The color of the links indicated if an association represent a positive (blue) or negative (red) correlation between the pathways. Each link includes an association strength value, which is a measure of the degree of relationship between the genes that belong two the linked pathways.
The node boxes include a plot that represents the pathway behavior along the time. Each node is also linked to its corresponding KEGG pathway diagram, where the most relevant genes of the signature were painted using the Paintomics tool. The pathway graph can be visualized clicking on the node.
These relevant genes, named as driving genes in PANA, represent the variables with major contribution to the conformation and variability of the pathway signatures. The highlight of these genes was carried out in order to facilitate the analysis and interpretation of the pathway interactions. The driving genes can be accessed from the pathway diagrams by clicking in the painted gene boxes.
For a particular set of links, interpretations of the pathway interactions were introduced. These links are denoted with thicker arrows. The pdf files with these explanations can be accessed by clicking on these pathway links.
Additional data about YCCPN links, like the bASp and mASp values, the number of genes of the linked pathways, driving genes and other relevant information is included in the following table. Another information, useful for the network analysis, is given in the following link.