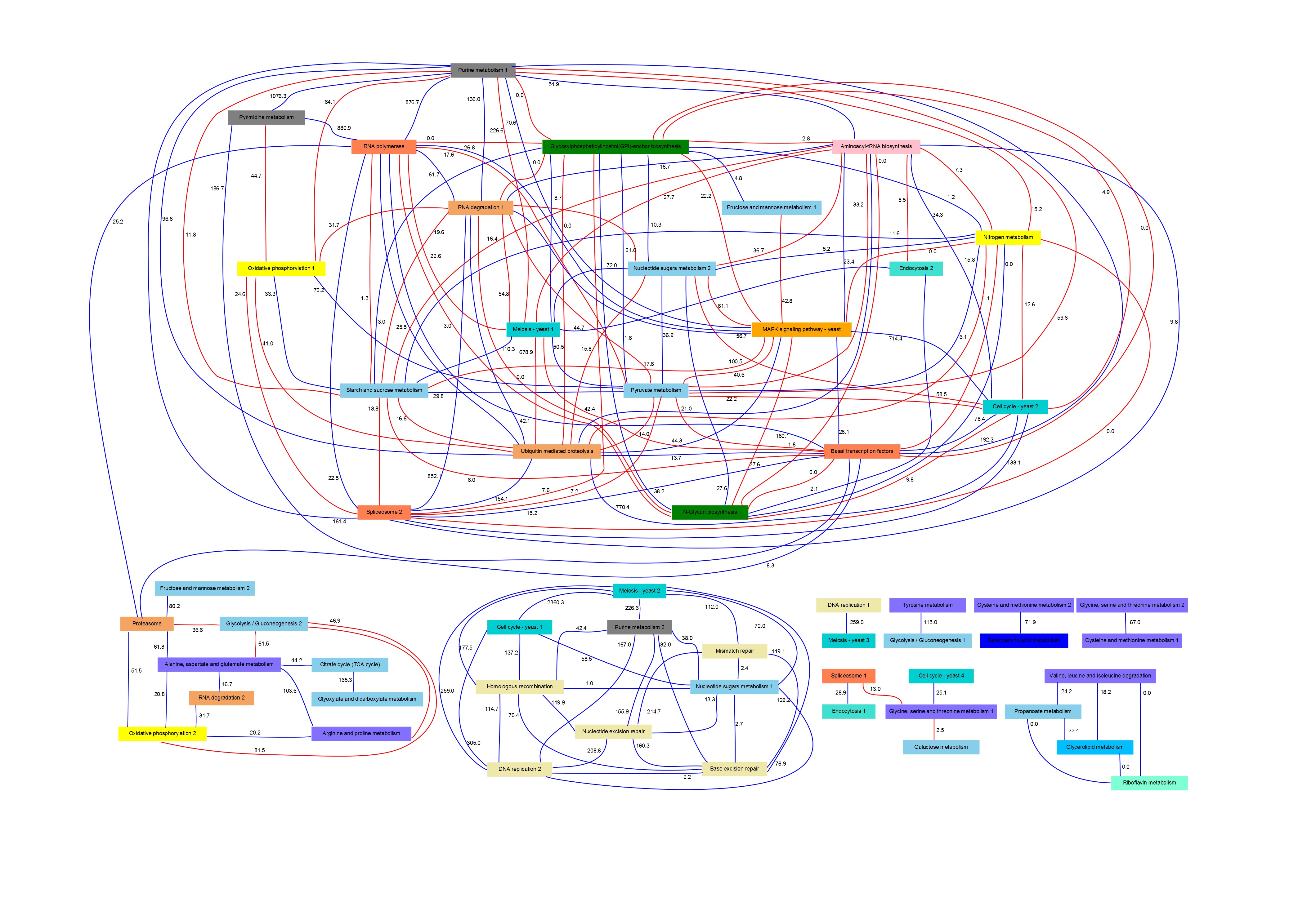
Association Strength Values
In order to evaluate the functional significance of the pathway networks, a gene functional annotation data contained in YestNet2 (Lee et al., 2007) was used. This database describes 102,803 functional associations among 5,483 yeast genes obtained from different experimental evidences and provides a association score for each pair-wise gene relationship.
In PANA, given a pair of pathways, functional association strength between these pathways is computed as the sum of the functional association scores of all gene pairs that can be established between these pathways. In this way, an independent association strength value between each pair of yeast pathways was obtained for validation purposes.
Therefore, in mathematical terms, the association strength value that corresponds to the link between two pathways i and j, ASpij, can be computed as follows:
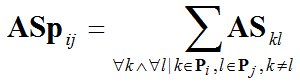
Where:
ASkl is the association score between genes k and l, obtained from YeastNet2, and
Pi and Pj are the sets of genes included in pathways i and j respectively.
The ASp values associated to the YCCPN links are depicted over the network edges (see).