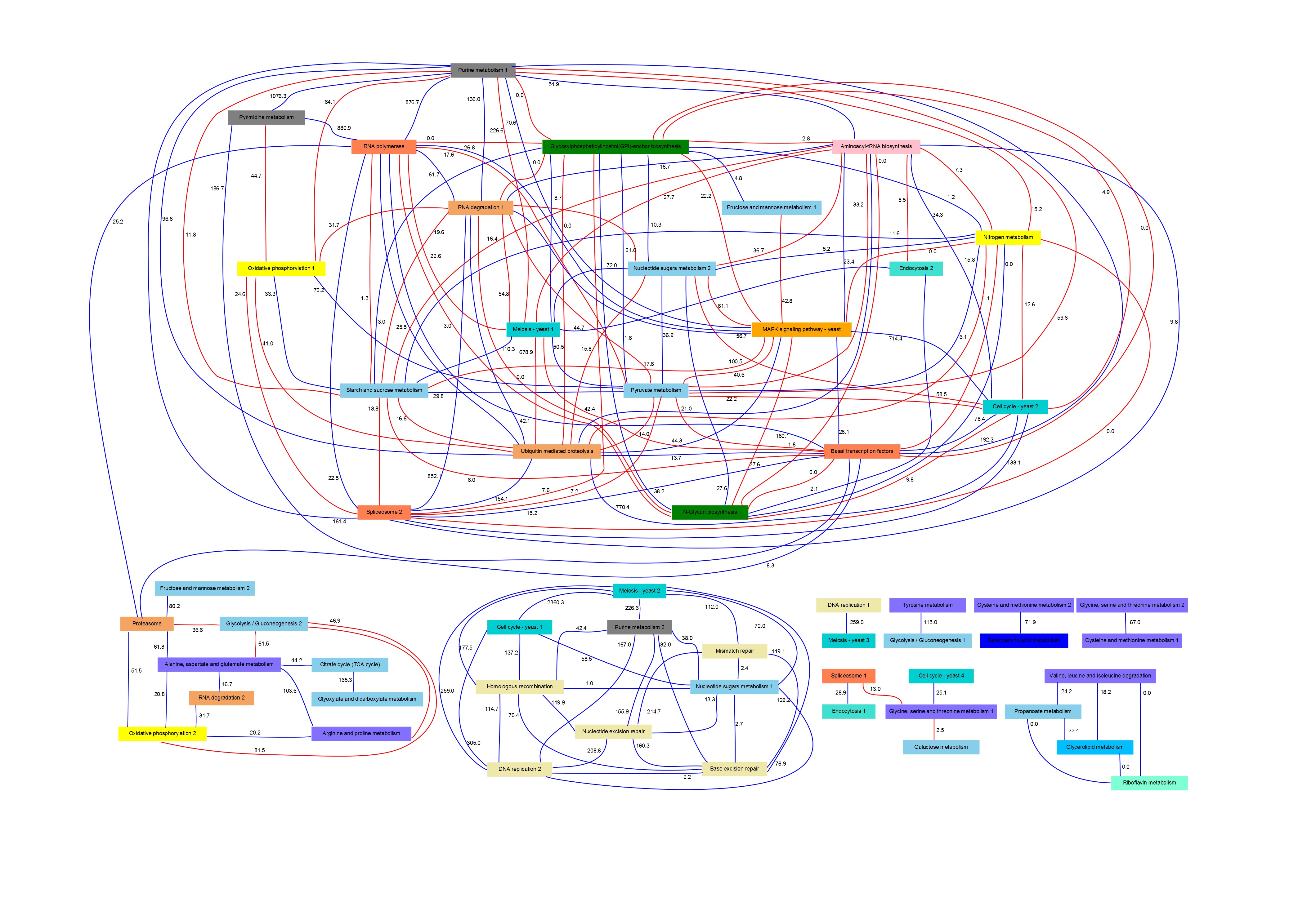
Pathway Signatures
PANA is a tool for the inference of associations at pathway level. In short, this method tries to capture the dynamics of the pathway behavior from the temporal expression profiles of their genes using principal component analysis (PCA). In this way, the temporal profile of a pathway is modeled by a principal component obtained by PCA, called pathway signature, whose score values represent a global measure of gene expression variations of the pathway genes along the sampled time-points.
It is important to mention that a network may include more than one signature per pathway. The quantity of selected signatures depends on the variability of the gene expression values in the data and a cutoff parameter value chosen by the user. If more than one signature is selected for a pathway, this situation is indicated in the network nodes adding consecutive numbers to the pathway name.
In this pdf file three plots are included for each pathway: the pathway signature profile, the loading value curve (with the identification of the gene with higher loading value), and the temporal profile that corresponds to this gene. This information is useful for understanding the direction of the principal component that represents the signature pathway.