PANA
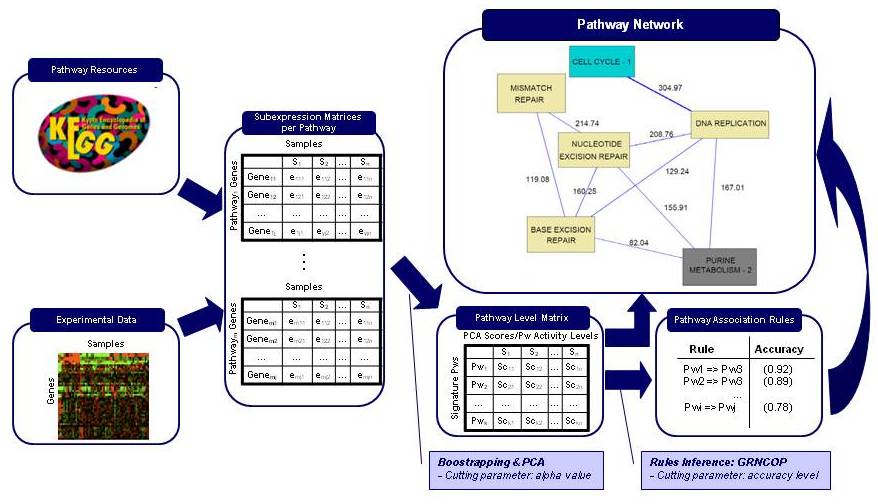
PANA (PAthway Network Analysis) is a novel approach for data analysis in Systems Biology. This method enables the definition of networks of relationships between molecular pathways and permits the study of their changes across experimental situations. The methodology has been developed for the analysis of gene expression data but it can be easily extended to consider other types of molecular high-throughput measurements.
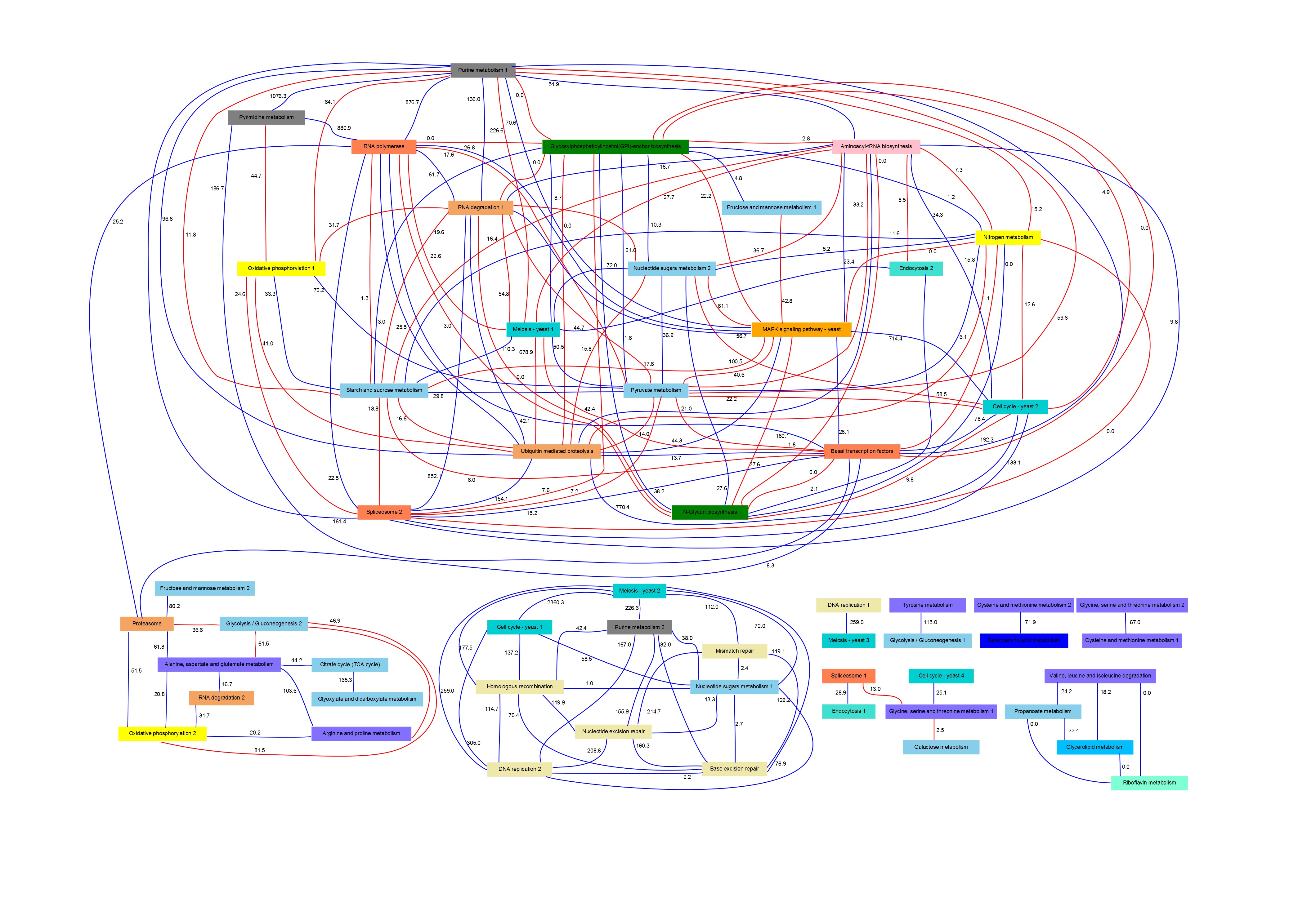
PANA combines dimensionality reduction methods with machine learning techniques for the inference of association rules. Given a gene expression experiment and a functional annotation of genes into pathways or functional modules, the method uses principal component analysis (PCA) to obtain for each pathway a variation signature that compresses the gene expression data for the gene members of the pathway. Next step consists in applying an adaptive method for extracting association rules based on mutual information maximization (Ponzoni et al. 2007). This method derives relationships between the pathway signatures. These relationships can be represented in the form of a network of pathway interactions where direct and inverse links can be displayed.
In particular, in this page we present results obtained for Saccharomyces Cerevisiae cell cycle time series.